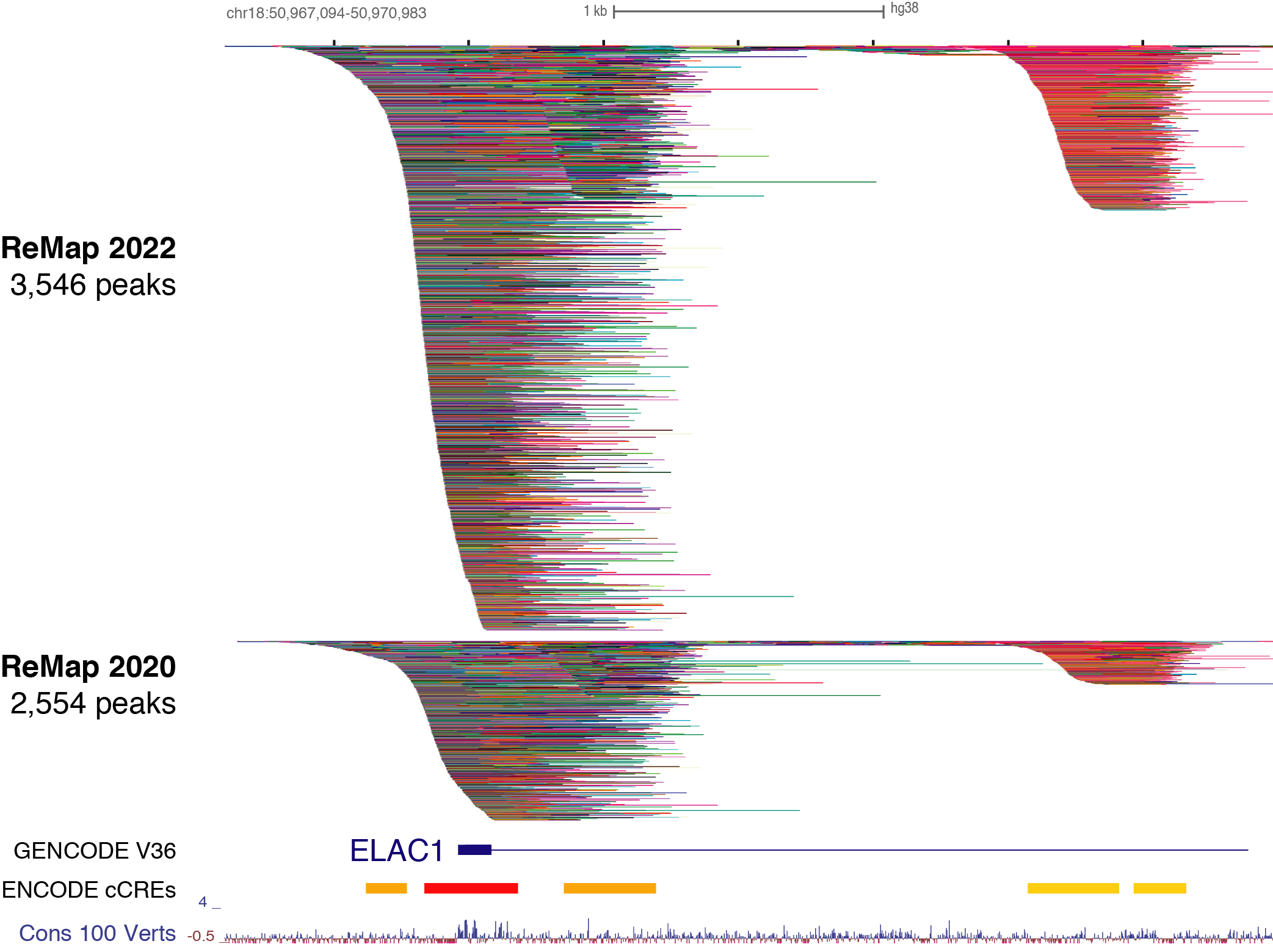
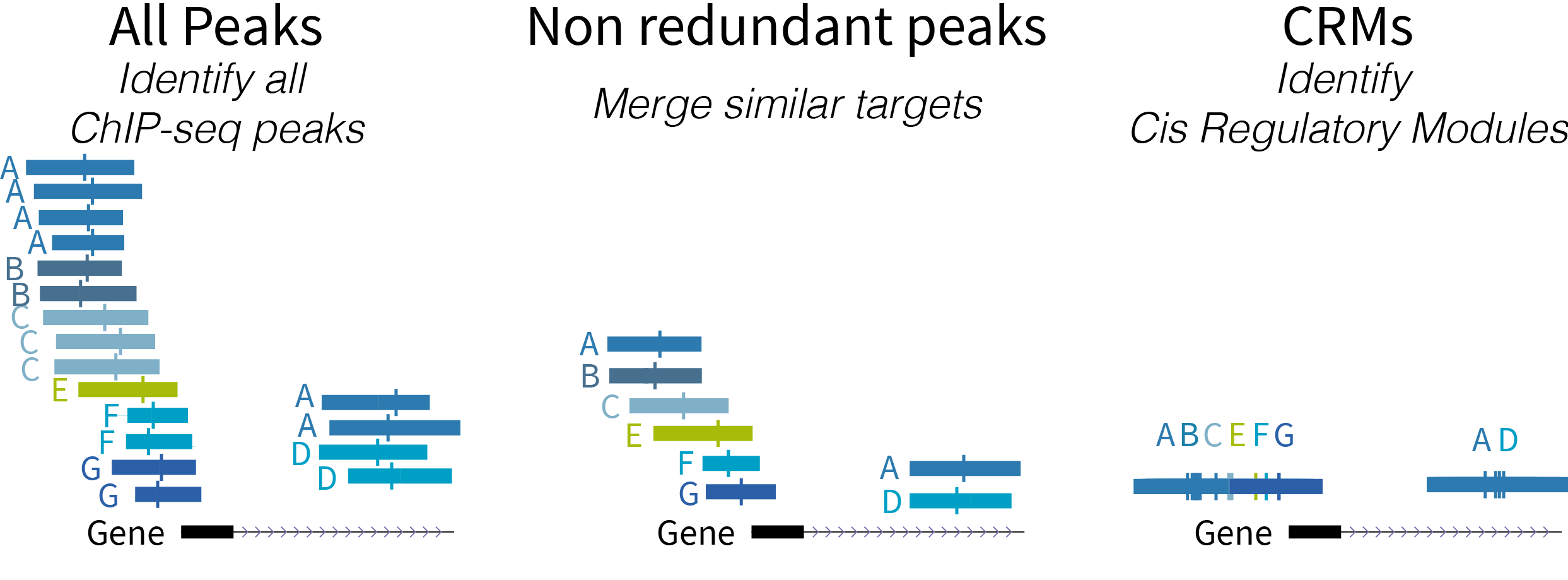
With ENCODE from UCSCC
Perl Script Explanation: Fetching cCREs and ReMap Peaks
This page explains the Perl script which fetches cCREs from UCSC and retrieves ReMap peaks within those cCRE regions.
1. Setup and Initialization
First, we initialize the necessary Perl modules for HTTP requests and JSON handling:
use strict;
use warnings;
use LWP::UserAgent;
use JSON;
The script makes HTTP requests to APIs, so it uses the LWP::UserAgent module. JSON responses are parsed using the JSON module.
2. Defining the UserAgent
We define a UserAgent object which will handle the HTTP requests and set a timeout of 10 seconds. Additionally, SSL verification is disabled for testing purposes:
my $ua = LWP::UserAgent->new;
$ua->timeout(10);
$ua->ssl_opts(verify_hostname => 0, SSL_verify_mode => 0x00);
This is necessary to avoid SSL errors when connecting to the UCSC API over HTTPS.
3. Fetching cCREs from UCSC
The script fetches cCRE (candidate Cis-Regulatory Elements) data from the UCSC Genome Browser's API:
my $ucsc_url = "https://api.genome.ucsc.edu/getData/track?genome=hg38;track=encodeCcreCombined;chrom=chr1;start=1470000;end=1480000";
my $response = $ua->get($ucsc_url);
The URL is specifically designed to fetch cCRE data for chromosome 1 between positions 1,470,000 and 1,480,000. The response from this request will be in JSON format.
4. Handling the Response and Parsing JSON
If the request is successful, the JSON response is parsed, and the script checks if it contains the required cCRE data:
if ($response->is_success) {
my $content = $response->decoded_content;
my $ccres_json = decode_json($content);
unless (exists $ccres_json->{encodeCcreCombined} && @{$ccres_json->{encodeCcreCombined}}) {
die "No cCREs found in the response.";
}
The JSON structure is decoded into a Perl hash, and the script checks if the key encodeCcreCombined exists, which holds the cCRE data.
5. Extracting cCRE Information
For each cCRE element in the response, the script extracts relevant fields such as chromosome, start, end, ucscLabel, and description. This information is printed to the console:
foreach my $ccre (@{$ccres_json->{encodeCcreCombined}}) {
my $chrom = $ccre->{chrom};
my $start = $ccre->{chromStart};
my $end = $ccre->{chromEnd};
my $ucsc_label = $ccre->{ucscLabel};
my $description = $ccre->{description};
print "\n\ncCRE: Chromosome: $chrom, Start: $start, End: $end\n";
print "UCSC Label: $ucsc_label\n";
print "Description: $description\n";
}
Each cCRE is printed with its coordinates, UCSC label, and description.
6. Fetching ReMap Peaks for Each cCRE
After extracting the cCRE data, the script constructs a URL to fetch ReMap peaks within each cCRE region:
my $remap_url = "http://localhost:8000/api/V1/get_peaks/2022/hg38/all/$chrom:$start-$end";
my $remap_response = $ua->get($remap_url);
This fetches the ReMap peaks data that overlap with the current cCRE. The start and end positions are dynamically set based on the cCRE coordinates.
7. Parsing and Formatting ReMap Peaks
For each ReMap peak, the script extracts fields such as chromosome, start, end, experiment, transcription factor (TF), and biotype. These are printed in BED format:
foreach my $peak (@{$remap_json->{peaks}}) {
my $peak_chrom = $peak->{peakValues}{chrom};
my $peak_start = $peak->{peakValues}{chromStart};
my $peak_end = $peak->{peakValues}{chromEnd};
my $experiment = $peak->{peakValues}{name}{data}{Experiment};
my $tf = $peak->{peakValues}{name}{data}{TF};
my $biotype = $peak->{peakValues}{name}{data}{Biotype};
# Create BED format
my $name = join(":", $experiment, $tf, $biotype);
my $score = 1000;
my $strand = ".";
print "$peak_chrom\t$peak_start\t$peak_end\t$name\t$score\t$strand\n";
}
The output is printed in the following BED format: chrom, start, end, name, score, and strand. This is the standard format used for genomic data files.
#!/usr/bin/perl
use strict;
use warnings;
use LWP::UserAgent;
use JSON;
my $verbose = 0;
# Initialize UserAgent for HTTP requests
my $ua = LWP::UserAgent->new;
$ua->timeout(10);
# Disable SSL verification (for testing purposes)
$ua->ssl_opts(verify_hostname => 0, SSL_verify_mode => 0x00);
# UCSC API endpoint for cCREs
my $ucsc_url = "https://api.genome.ucsc.edu/getData/track?genome=hg38;track=encodeCcreCombined;chrom=chr1;start=1470000;end=1480000";
# Fetch cCREs from UCSC
my $response = $ua->get($ucsc_url);
if ($response->is_success) {
my $content = $response->decoded_content;
my $ccres_json = decode_json($content);
# Print cCREs data for debugging
print "UCSC cCRE Data:\n" if $verbose;
print to_json($ccres_json, { pretty => 1 }) if $verbose;
# Check if cCREs data is available
unless (exists $ccres_json->{encodeCcreCombined} && @{$ccres_json->{encodeCcreCombined}}) {
die "No cCREs found in the response.";
}
# Loop through each cCRE and fetch ReMap peaks within this region
foreach my $ccre (@{$ccres_json->{encodeCcreCombined}}) {
my $chrom = $ccre->{chrom};
my $start = $ccre->{chromStart};
my $end = $ccre->{chromEnd};
my $ucsc_label = $ccre->{ucscLabel};
my $description = $ccre->{description};
# Print extracted values along with ucscLabel and description
print "\n\ncCRE: Chromosome: $chrom, Start: $start, End: $end\n";
print "UCSC Label: $ucsc_label\n";
print "Description: $description\n";
# Prepare ReMap API URL
my $remap_url = "http://localhost:8000/api/V1/get_peaks/2022/hg38/all/$chrom:$start-$end";
print "ReMap API URL: $remap_url\n"; # Debug URL
# Fetch ReMap peaks within cCRE
my $remap_response = $ua->get($remap_url);
if ($remap_response->is_success) {
my $remap_content = $remap_response->decoded_content;
my $remap_json = decode_json($remap_content);
# Print fetched ReMap peaks for debugging
print "ReMap Peaks Data:\n" if $verbose;
print to_json($remap_json, { pretty => 1 }) if $verbose;
# Check if ReMap peaks data is available
unless (exists $remap_json->{peaks} && @{$remap_json->{peaks}}) {
warn "No peaks found in the ReMap response for cCRE: $chrom:$start-$end";
next;
}
# Loop through each ReMap peak within the cCRE region
foreach my $peak (@{$remap_json->{peaks}}) {
my $peak_chrom = $peak->{peakValues}{chrom};
my $peak_start = $peak->{peakValues}{chromStart};
my $peak_end = $peak->{peakValues}{chromEnd};
my $experiment = $peak->{peakValues}{name}{data}{Experiment};
my $tf = $peak->{peakValues}{name}{data}{TF};
my $biotype = $peak->{peakValues}{name}{data}{Biotype};
# Create BED format (chrom, start, end, name, score, strand)
my $name = join(":", $experiment, $tf, $biotype);
my $score = "."; # Default score (can be adjusted based on your needs)
my $strand = "."; # Strand information (set as '.' if unknown)
# Print the BED formatted line
print "$peak_chrom\t$peak_start\t$peak_end\t$name\t$score\t$strand\n";
}
} else {
warn "Failed to fetch ReMap data: " . $remap_response->status_line;
}
}
} else {
die "Failed to fetch UCSC cCRE data: " . $response->status_line;
}
Benchmarks by number of regions
In our benchmark section, we put our API through its paces by gradually ramping up the number of genomic regions queried. This hands-on testing helps us understand how our API performs under different workloads, giving scientists and bioinformaticians valuable insights into its speed and reliability across various scenarios. By simulating real-world usage, we ensure that our API can handle the demands of large-scale genomic analyses, empowering researchers with the tools they need for their studies.
Advise
At the light of our benchmark, we stronlgy advise to query our catalogue by batch of maximum XXXX coordinates.
Benchmarks by size of regions
In another part of our benchmarking, we explore the API's performance as we increase the size of the genomic regions being queried. This analysis helps us understand how the API handles larger genomic segments, offering scientists and bioinformaticians practical insights into its scalability and efficiency. By simulating different genomic contexts, we ensure that our API remains reliable and responsive, even when dealing with extensive genomic data sets, thus providing researchers with robust tools for their analyses.
Advise
At the light of our benchmark, we stronlgy advise to query our catalogue with cooridnates no larger than XX Kb / 0.X Mb as it will impact the speed or your query. Query many shorter regions will be fatser one large region.
Benchmark conclusion
Vivamus efficitur fringilla ullamcorper. Cras condimentum condimentum mauris, vitae facilisis leo. Aliquam sagittis purus nisi, at commodo augue convallis id. Sed interdum turpis quis felis bibendum imperdiet. Mauris pellentesque urna eu leo gravida iaculis. In fringilla odio in felis ultricies porttitor. Donec at purus libero. Vestibulum libero orci, commodo nec arcu sit amet, commodo sollicitudin est. Vestibulum ultricies malesuada tempor.
Licensing
The ReMap catalogues (2022, 2020, 2018, 2015) are under CC BY-NC 4.0 international license, while ReMap-REST, ReMapEnrich, ReMap-Pipeline, are under GNU GPLv3 licence.
ReMap is a database of transcriptional regulators peaks derived from curated ChIP-seq, ChIP-exo, DAP-seq experiments in Human, Mouse, Fruit Fly and Arabidopsis Thaliana.
The current ReMap version is the 2022 (4rth) release.